Introduction to point
This project was originally inspired by the lack of detailed insight in the inner workings of the default software for the Cameca NanoSIMS50L (Utrecht University). Hence this project has the objective of processing raw ion count data into ion and isotope ratios of point-sourced measurements. Combined with statistics for the internal and external precision of, respectively, individual analyses and complete series of analyses, this functionality allows for the interrogation of the analytical consistency. Access to raw ion count data is, furthermore, useful as it allows detection of anomalous values associated with e.g. machine instability or heterogeneity of the analysed substrate. Upon detection, anomalous values can be omitted or further analysed to delineate the source of variation.
The point package is still under development but the master branch is functioning. Functionality is automatically tested with CI.
Credits
The construction of the R (R Core Team 2022) package point and associated documentation was aided by the packages; devtools (Wickham, Hester, et al. 2021), roxygen2 (Wickham, Danenberg, et al. 2021), testthat (Wickham 2022), knitr (Xie 2014 ; Xie 2015), rmarkdown (Xie, Allaire, and Grolemund 2018; Xie, Dervieux, and Riederer 2020), bookdown (Xie 2016) and the superb guidance in the book: R packages: organize, test, document, and share your code, by Wickham (2015). In addition, this package relies on a set of external packages from the tidyverse universe, including: dplyr (Wickham et al. 2022), tidyr (Wickham and Girlich 2022), tibble (Müller and Wickham 2021), stringr (Wickham 2019), readr (Wickham, Hester, and Bryan 2022), magrittr (Bache and Wickham 2022), rlang (Henry and Wickham 2022), and purrr (Henry and Wickham 2020) for internal functioning as well as specialised statistics; polyaAeppli (Burden 2014), broom (Robinson, Hayes, and Couch 2022), broom.mixed (Bolker and Robinson 2022), nlme (Pinheiro, Bates, and R Core Team 2022), and mgcv (Wood 2022). Plots are made with ggplot2 (Wickham 2016)
Installation
You can install the released version of point
# Install point from GitHub:
# install.packages("devtools")
devtools::install_github("MartinSchobben/point")The point workflow
A more detailed outline of the general point workflow is given in the vignette IC-introduction (vignette("IC-introduction")).
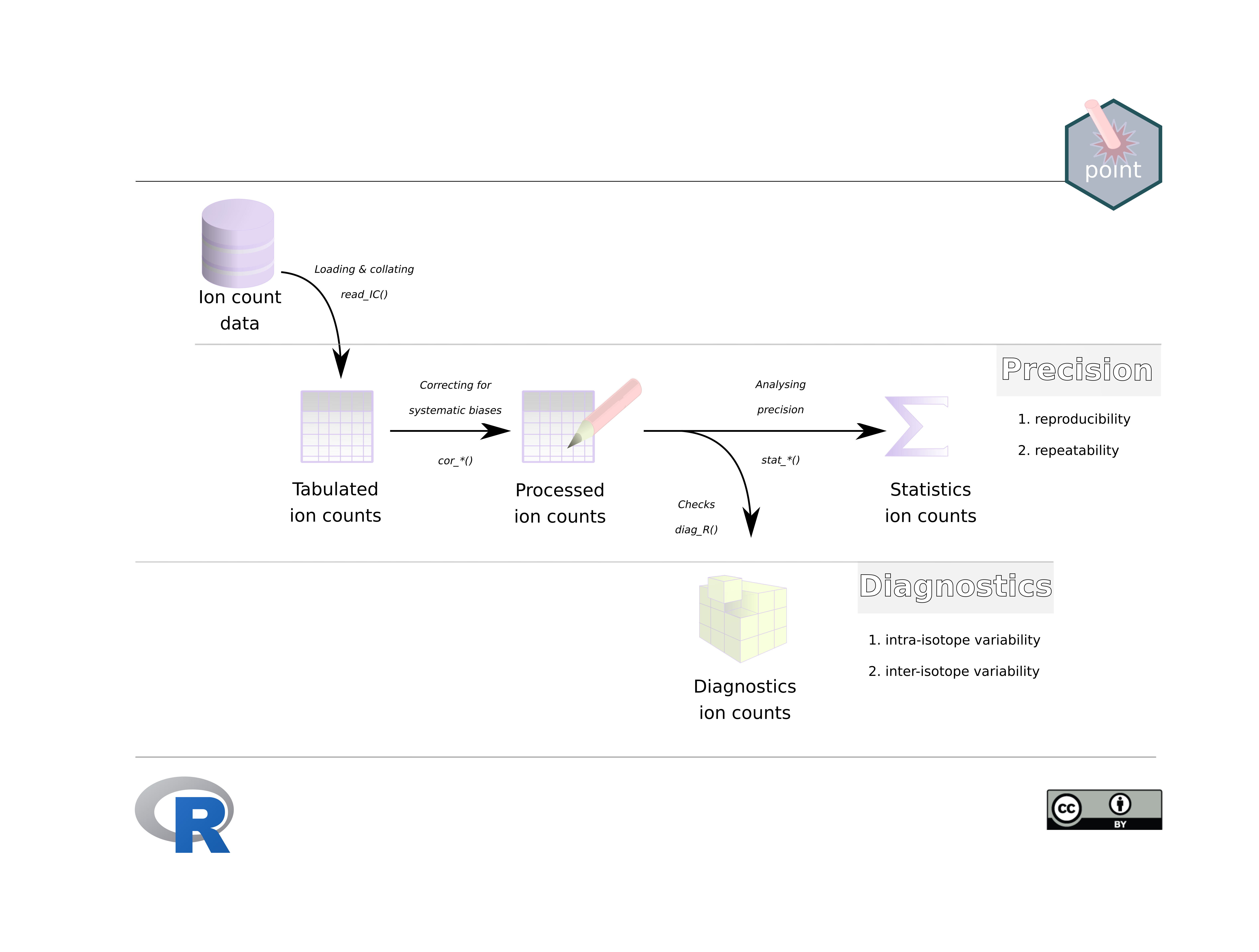
To read, process and analyse raw ion count data use the functions:
Example 1: internal precision of isotope ratios
This is an example of how Cameca NanoSIMS50L raw data files can be extracted, processed and analysed for the 13C/12C isotope ratio (). This produces a tibble with descriptive and predictive (Poisson) statistics (demarcated with an ^) of the ion count data. This can be done for single analysis in order to obtain internal precision.
# Use point_example() to access the examples bundled with this package in the
# inst/extdata directory.
# Raw data containing 13C and 12C counts on carbonate
tb_rw <- read_IC(point_example("2018-01-19-GLENDON"), meta = TRUE)
#> Registered S3 methods overwritten by 'readr':
#> method from
#> as.data.frame.spec_tbl_df vroom
#> as_tibble.spec_tbl_df vroom
#> format.col_spec vroom
#> print.col_spec vroom
#> print.collector vroom
#> print.date_names vroom
#> print.locale vroom
#> str.col_spec vroom
# Processing raw ion count data
tb_pr <- cor_IC(tb_rw)
# Internal precision for 13C/12C ratios
tb_R <- stat_R(tb_pr, "13C", "12C", sample.nm, file.nm, .label = "webtex")| sample.nm | file.nm | ratio.nm |
|
|
|
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Belemnite,Indium | 2018-01-19-GLENDON_1_1 | 13C/12C | 3900 | 0.011 | 0.001021 | 93.0 | 1.63e-05 | 1.49 | 0.001019 | 92.8 | 1.63e-05 | 1.49 | 1.00 |
| Belemnite,Indium | 2018-01-19-GLENDON_1_2 | 13C/12C | 3900 | 0.011 | 0.000779 | 70.8 | 1.25e-05 | 1.13 | 0.000771 | 70.1 | 1.23e-05 | 1.12 | 1.02 |
| Belemnite,Indium | 2018-01-19-GLENDON_1_3 | 13C/12C | 3900 | 0.011 | 0.000733 | 66.5 | 1.17e-05 | 1.06 | 0.000722 | 65.5 | 1.16e-05 | 1.05 | 1.03 |
Example 2: external precision of isotope ratios
To calculate the external reproducibility of isotope ratios one needs to use the .nest argument to define sets of analyses.
# external precision for 13C/12C ratios
tb_R <- stat_R(tb_pr, "13C", "12C", sample.nm, file.nm, .nest = file.nm,
.label = "webtex")| sample.nm | ratio.nm |
|
|
|
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Belemnite,Indium | 13C/12C | 3 | 0.011 | 2.03e-05 | 1.85 | 1.17e-05 | 1.07 | 1.3e-05 | 1.18 | 7.5e-06 | 0.681 | 2.45 |
For more detailed information:
IC-read (vignette("IC-read")): reading raw ion count data use
IC-process (vignette("IC-process")): processing ion count data
IC-precision (vignette("IC-precision")): statistics concerning ion count precision
IC-diagnostics (vignette("IC-diagnostics")): diagnostics on internal variation
References
Bache, Stefan Milton, and Hadley Wickham. 2022. Magrittr: A Forward-Pipe Operator for r. https://CRAN.R-project.org/package=magrittr.
Bolker, Ben, and David Robinson. 2022. Broom.mixed: Tidying Methods for Mixed Models. https://github.com/bbolker/broom.mixed.
Burden, Conrad. 2014. polyaAeppli: Implementation of the Polya-Aeppli Distribution. https://CRAN.R-project.org/package=polyaAeppli.
Henry, Lionel, and Hadley Wickham. 2020. Purrr: Functional Programming Tools. https://CRAN.R-project.org/package=purrr.
———. 2022. Rlang: Functions for Base Types and Core r and Tidyverse Features. https://CRAN.R-project.org/package=rlang.
Müller, Kirill, and Hadley Wickham. 2021. Tibble: Simple Data Frames. https://CRAN.R-project.org/package=tibble.
Pinheiro, José, Douglas Bates, and R Core Team. 2022. Nlme: Linear and Nonlinear Mixed Effects Models. https://svn.r-project.org/R-packages/trunk/nlme/.
R Core Team. 2022. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
Robinson, David, Alex Hayes, and Simon Couch. 2022. Broom: Convert Statistical Objects into Tidy Tibbles. https://CRAN.R-project.org/package=broom.
Wickham, Hadley. 2015. R Packages: Organize, Test, Document, and Share Your Code. O’Reilly Media, Inc. https://r-pkgs.org/.
———. 2016. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. https://ggplot2.tidyverse.org.
———. 2019. Stringr: Simple, Consistent Wrappers for Common String Operations. https://CRAN.R-project.org/package=stringr.
———. 2022. Testthat: Unit Testing for r. https://CRAN.R-project.org/package=testthat.
Wickham, Hadley, Peter Danenberg, Gábor Csárdi, and Manuel Eugster. 2021. Roxygen2: In-Line Documentation for r. https://CRAN.R-project.org/package=roxygen2.
Wickham, Hadley, Romain François, Lionel Henry, and Kirill Müller. 2022. Dplyr: A Grammar of Data Manipulation. https://CRAN.R-project.org/package=dplyr.
Wickham, Hadley, and Maximilian Girlich. 2022. Tidyr: Tidy Messy Data. https://CRAN.R-project.org/package=tidyr.
Wickham, Hadley, Jim Hester, and Jennifer Bryan. 2022. Readr: Read Rectangular Text Data. https://CRAN.R-project.org/package=readr.
Wickham, Hadley, Jim Hester, Winston Chang, and Jennifer Bryan. 2021. Devtools: Tools to Make Developing r Packages Easier. https://CRAN.R-project.org/package=devtools.
Wood, Simon. 2022. Mgcv: Mixed GAM Computation Vehicle with Automatic Smoothness Estimation. https://CRAN.R-project.org/package=mgcv.
Xie, Yihui. 2014. “Knitr: A Comprehensive Tool for Reproducible Research in R.” In Implementing Reproducible Computational Research, edited by Victoria Stodden, Friedrich Leisch, and Roger D. Peng. Chapman; Hall/CRC. http://www.crcpress.com/product/isbn/9781466561595.
———. 2015. Dynamic Documents with R and Knitr. 2nd ed. Boca Raton, Florida: Chapman; Hall/CRC. https://yihui.org/knitr/.
———. 2016. Bookdown: Authoring Books and Technical Documents with R Markdown. Boca Raton, Florida: Chapman; Hall/CRC. https://bookdown.org/yihui/bookdown.
Xie, Yihui, J. J. Allaire, and Garrett Grolemund. 2018. R Markdown: The Definitive Guide. Boca Raton, Florida: Chapman; Hall/CRC. https://bookdown.org/yihui/rmarkdown.
Xie, Yihui, Christophe Dervieux, and Emily Riederer. 2020. R Markdown Cookbook. Boca Raton, Florida: Chapman; Hall/CRC. https://bookdown.org/yihui/rmarkdown-cookbook.
