Reading matlab files
data.RmdThe R package pointapply contains the code and data to reconstruct the publication: Martin Schobben, Michiel Kienhuis, and Lubos Polerecky. 2021. New methods to detect isotopic heterogeneity with Secondary Ion Mass Spectrometry, preprint on Eartharxiv.
Introduction
The ion count data encompasses Secondary Ion Mass Spectrometry (SIMS) analyses by a Cameca NanoSIMS 50L (Utrecht University) on the certified reference standard MEX (Jochum and Brueckner 2008; Rollion-Bard, Mangin, and Champenois 2007), which is an sedimentary carbonate, and the (uncertified) crystalline calcite MON.
The following packages are used in the for the examples and layout of the document.
library(point) # regression diagnostics
library(pointapply) # this paperThe package point (Schobben 2022) is especially designed to perform the intra- and inter-analysis isotope tests highlighted in the publication.
Download data
The raw ion count data is generated with Look@NanoSIMS (Polerecky et al. 2012) as LANS and comes in matlab format. Raw data files should first be downloaded from the external Zenodo repository with the function download_point(). Consecutively, these raw data files can be transformed into the right format with the functions highlighted in this vignette.
# use download_point() to obtain processed data.
# this only has to be done once after installation
download_point(type = "raw")Extract ion count data
The two “real” datasets of Schobben, Kienhuis and Polerecky 2021
The two “real” ion mapping datasets (as opposed to simulated data) are included as external data (inst/extdata directory) in the package pointapply, and the pathways can be conveniently generated by the function get_matlab().
get_matlab("2020-08-20-GLENDON") # MEX Matlab files were generated by the nanoSIMS imaging software Look@NanoSIMS (Polerecky et al. 2012). This program contains algorithms for the alignment of pixels across successive depth, or depth-frames, of the ion mapping protocol.
The matlab-generated files contain multidimensional hyper-arrays (or ‘data cubes’), where ion count measurements have three spatial coordinates; width 256 pixels, height 256 pixels and 400 frames of depth, and can gave several bandwidths, which represent the different chemical species analysed by nanoSIMS. R.matlab (Bengtsson 2018) or readmat (???) can be used to load Matlab files, where the latter is the fastest method but relies on a full Matlab installation and thus a Matlab license.
# the files
MEX_files <- list.files(get_matlab("2020-08-20-GLENDON"), full.names = TRUE)
MON_files <- list.files(get_matlab("2020-08-28-GLENDON"), full.names = TRUE)
# read files (alternatively use R.matlab)
MEX <- lapply(MEX_files, readmat::read_mat)
MON <- lapply(MON_files, readmat::read_mat)
# get measured species names
MEX_species <- vapply(MEX, attr, character(1), "file")
MON_species <- vapply(MEX, attr, character(1), "file")
MEX_species <- gsub("_cnt", "", MEX_species)
MON_species <- gsub("_cnt", "", MON_species)
# set names
MEX <- unlist(MEX, recursive = FALSE) |> stats::setNames(MEX_species)
MON <- unlist(MON, recursive = FALSE) |> stats::setNames(MON_species)The grid_aggregate function is designed to aggregate over a specified dimension (e.g. "height" see example below) and a set of species (e.g. c("12C", "13C") see example below). The grid-cell size for aggregation can be set by the grid_cell argument, which should be an integer exponent with base two (64 pixels by 64 pixels in this example).
grid_aggregate(MEX, "height", 64, c("12C", "13C"))The tibble contains the following variables:
-
sample.nm: The sample name, as provided through thetitleargument of the function. -
file.nm: The original file name. -
grid_size.nm: Convert to pixels to grid-cell size (e.g. \(\mu\)m) dimensions, with the argumentsscalerwhich defaults to 0.16\(\,\mu\)m pixel-. -
grid.nm: Grid-cell number. -
dim.nm: The step value for the dimensional scale for aggregation in pixels (ifplane = heightorplane = width) or depth-frames (ifplane = depth). *dim_name.nm: The name for the dimensional scale for aggregation (e.g. height). -
species.nm: The chemical species name. -
N.rw: The ion counts. -
t.nm: The time increments (in seconds), which are based on \(1\,\)millisecond per pixel.
Grid-cell size
The grid-cell size has important consequences for the data structure of the ion count measurements, and this is further explored in the paper and the vignette Validation of regression assumptions (vignette("regression")). One can visualise the layout of the grid in a two dimensional representation, as follows (Supplementary Fig. 2). This representation could be said to resemble a flattened cardboard box.
# generate sketch of grid layout
gg_sketch(grid_cell = 64, save = TRUE) 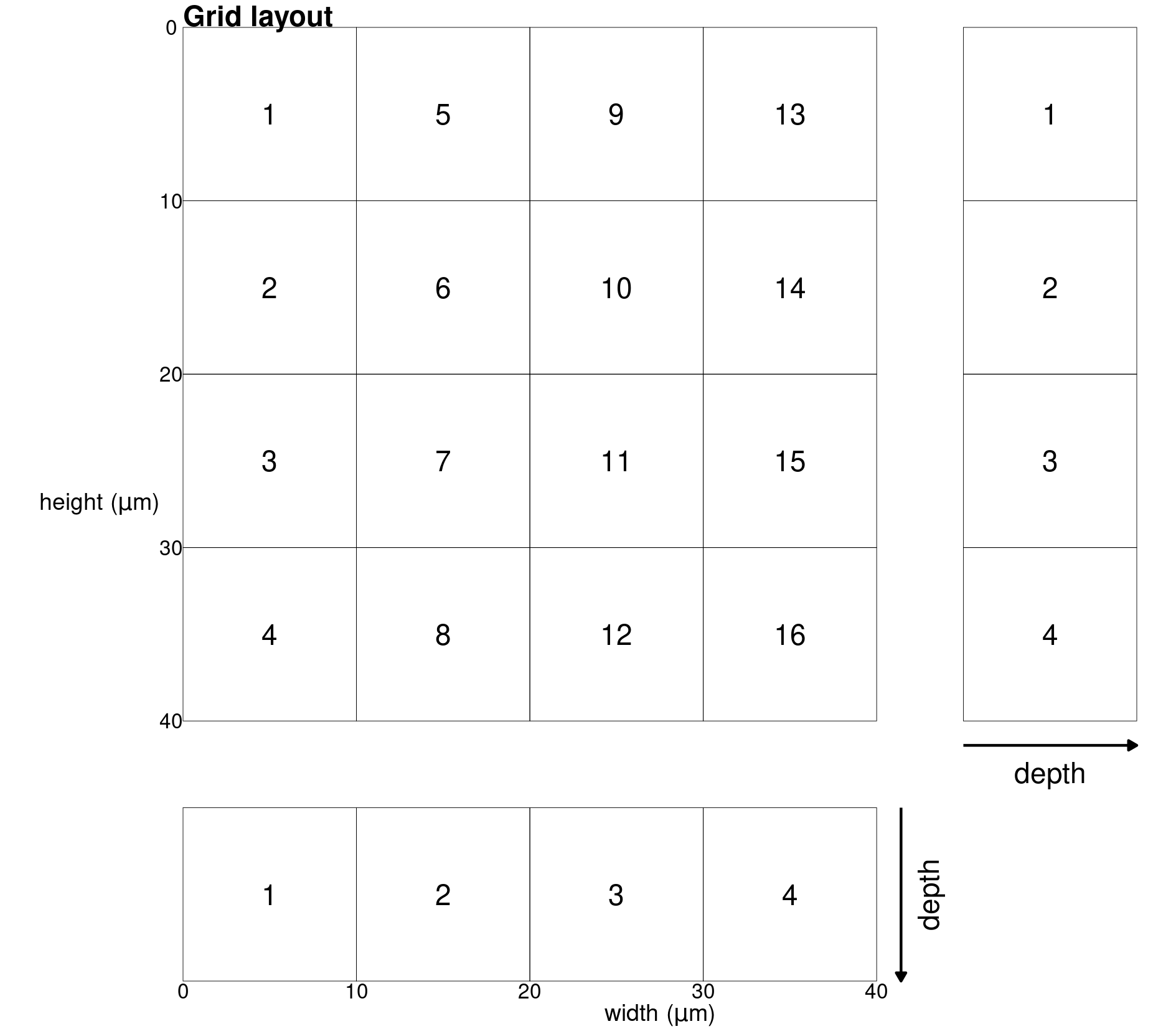
Sketch of the grid layout with grid-cell size of 64 pixels by 64 pixels.
Read data for different grid-cell sizes and planes
The following code was use to produce data aggregation for 12C and 13C over different grid-cell sizes. This was done to allow an assessment of the sensitivity of the intra- and inter- analysis isotope tests towards the internal data structure, where smaller dimensions cause the underlying structure to deviate from a Gaussian distribution (see vignette Validation of regression assumptions (vignette("regression")) and the original paper).
# 4, 8, 16, 32, 64, and 128 px grid-cell size for aggregating data
vc_px <- sapply(2:7, function(x) 2 ^ x)
# tuning the perfect grid dimensions (MEX)
# the ion counts are corrected for systematic bias (`corrected` = TRUE)
grid_aggregate(MEX, c("height", "width", "depth"), grid_cell = 64,
species = c("12C", "13C"), title = "MEX", name = "map_sum_grid",
corrected = TRUE, save = TRUE) |>
# `tune_grid` allows parallel computing by setting number of cores
tune_grid_cell(vc_px, 4)
# MON
grid_aggregate(MON, c("height", "width", "depth"), grid_cell = 64,
species = c("12C", "13C"), title = "MON", name = "map_sum_grid",
corrected = TRUE, save = TRUE) |>
# `tune_grid` allows parallel computing by setting number of cores
tune_grid_cell(vc_px, 4) Read data for raster images
By keeping the original raster we can also produce ion maps by aggregation of ion counts for each of the planes.
# raster images (MEX)
grid_aggregate(
MEX,
c("height", "width", "depth"),
title = "MEX",
name = "map_raster_image",
save = TRUE
)
# raster images (MON)
grid_aggregate(
MON,
c("height", "width", "depth"),
title = "MON",
name = "map_raster_image",
save = TRUE
)Read data for selected grid-cells
Finally, we can subset full ion count data cubes from selected grid-cells on a specified plane. Here grid-cells 2 and 9 were extracted on a 64 pixel by 64 pixel sized grid for MEX (see schematic figure above for orientation of grid-cells).
# full data-sets for grid-cell 2 and 9 of the depth plane (MEX)
grid_select(
MEX,
"depth",
grid_cell = 64,
select_cell = c(2, 9),
title = "MEX",
name = "map_full_grid",
save = TRUE
)References
Bengtsson, Henrik. 2018. R.matlab: Read and Write Mat Files and Call Matlab from Within R. https://github.com/HenrikBengtsson/R.matlab.
Jochum, Klaus Peter, and Stefanie M Brueckner. 2008. “Reference materials in geoanalytical and environmental research - Review for 2006 and 2007.” https://doi.org/10.1111/j.1751-908X.2008.00916.x.
Polerecky, Lubos, Birgit Adam, Jana Milucka, Niculina Musat, Tomas Vagner, and Marcel M. M. Kuypers. 2012. “Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology.” Environmental Microbiology 14 (4): 1009–23. https://doi.org/10.1111/j.1462-2920.2011.02681.x.
Rollion-Bard, Claire, Denis Mangin, and Michel Champenois. 2007. “Development and application of oxygen and carbon isotopic measurements of biogenic carbonates by ion microprobe.” Geostandards and Geoanalytical Research 31 (1): 39–50. https://doi.org/10.1111/j.1751-908X.2007.00834.x.
Schobben, Martin. 2022. Point: Reading, Processing, and Analysing Raw Ion Count Data. https://martinschobben.github.io/point/.